LAB5REP - Identification and Quantitative Analysis of a Monoprotic Acid by a Potentiometric PDF

| Title | LAB5REP - Identification and Quantitative Analysis of a Monoprotic Acid by a Potentiometric |
|---|---|
| Course | Quantitat Analytical Chem Lab |
| Institution | University of Georgia |
| Pages | 10 |
| File Size | 549.3 KB |
| File Type | |
| Total Downloads | 93 |
| Total Views | 182 |
Summary
Identification and Quantitative Analysis of a Monoprotic Acid by a
Potentiometric Titration...
Description
Identification and Quantitative Analysis of a Monoprotic Acid by a Potentiometric Titration Makaela Everett October 27, 2020 CHEM2300L – Wednesday A
Abstract The experiment completed was the identification and quantitative analysis of a monoprotic acid by a potentiometric titration. This was completed to understand the use of potentiometric titrations, to determine the molar mass of an unknown weak monoprotic acid, to determine the pKa values for an unknown weak monoprotic acid, and to confirm the identity of the unknown monoprotic acid through titration data collected . One goal was using potentiometric titration to help predict an unknown acid with titrations of a strong base, in the case of this experiment sodium hydroxide (NaOH) was used. The instrumentation for the potentiometric titration is studied with particular emphasis on automatic apparatus developments tending to simplify the use of the potentiometric method. The MicroLab instrument, capable of automatically performing and plotting potentiometric titrations, and adaptable to a variety of electrode systems, is used and data obtained is observed throughout the experiment. MicroLab is operated by calibrating the instrument as well as placing the electrode in the solution, while being able to plot the titration. The CurtiPlot was also used to help create a smooth titration curve and provide information about the titrations. Sample 9MA21 of the unknown acid was used for the experiment and based on the average molecular weight calculation of 144.8 g/mols, the standard deviation was 34.02 g/mol, and the percent relative standard deviation was 23.49%. The pKa’s average calculation of 3.286, the standard deviation was 0.1236, the percent relative standard deviation was 3.895%. The 95% confidence limit was ±38.50 g/mol for molar mass, and was ±0.1448 for pKa. It is assumed that maleic acid was used during the experiment as the unknown monoprotic acid.
Introduction For this experiment the student learned about the process of identification and quantitative analysis of a monoprotic acid by a potentiometric titration. Potentiometric titration usually takes place in the presence of a neutral electrolyte with a common counterion (2). The pKa is an experimentally determined parameter that tells us how tightly protons are bound to different compounds (3). The pKa measures how tightly a proton is held by a Bronsted acid, the higher the pKa of a Bronsted acid, the more tightly the proton is held, and the less easily the proton is given up (3). And monoprotic acid is defined as an acid that can only accept one proton (1). Indicators are used to help one see when the reaction has reached its endpoint, the indicators used in this experiment are phenolphthalein, bromothymol blue, and bromocresol purple. The phenolphthalein indicator is colorless in acidic solution and pink in basic solution. The bromothymol blue indication will be yellow in an acidic solution and blue in a basic solution. In addition, the bromocresol purple indicator is yellow in an acidic solution and purple in a basic solution. By using potentiometric titration, the MicroLab plotted the volume versus pH to observe the equivalence points in the graphs. Because the equilibrium of the analytical reaction is favorable, and the right combination of electrodes were chosen there was a sharp change in voltage at the equivalence point. The equivalence point is the slope of the titration curve is at a maximum. In other words, the rate of change of pH with addition of titrant is at its highest at the equivalence point. So, by plotting the rate of change of pH with change in volume added an endpoint can be found. This is also done with CuritPlot to create a smooth titration cure from the raw data and to help find the true endpoint of the titration. This is important in a chemistry laboratory because it shows when a reaction is completed or has gone to the other side of the reaction. Potentiometric titrations are important because when this method is precise a sample will give similar results and the significant difference will be low for the volume of what was titrated and the pKa found of what was titrated. Precision characteristics of the titration matter, as well as sensitivity, method stability and the tools used for calculating titration curves. This can be used to ensure that your method and instruments produces similar results and values. The purpose of this experiment was to identify a monoprotic acid by a potentiometric titration through three titration trials. And to find the molar mass and pKa values from the data retrieved from these titrations. The results from this will show how indicators have different pH
2
ranges in which a color change will occur, is present, and that titrations and titration curves follow procedure and that the calculations are properly performed. In order to analyze the moles, pKa, and the molar mass of the unknown acid, three titrations will be performed, and the values will be calculated using stoichiometry and other conversion factors. The total volume used to reach the endpoint of the titration will also display the mole concentration of the NaOH and the moles of the unknown acid. These values will later be used to calculate the molar mass of the unknown.
Procedure The procedure was carried out according to the experiment handout, Experiment 16: Identification and Quantitative Analysis of a Monoprotic Acid by a Potentiometric Titration
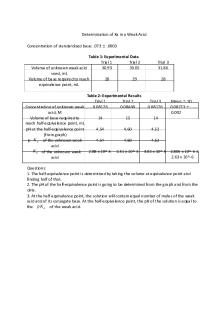
Results The results for the potentiometric titration of an unknown acid are shown in Table 1 below. The table details the mass of unknown acid used in grams. Along with the volume of NaOH used for the color change to occur, the volume of NaOH used for endpoint to be reached, the moles of NaOH. The moles and of NaOH were found by using Equation 1. The values for moles of NaOH were found to be 0.001096, 0.0006016, and 0.0006357. The table also displays the indicator used for each trial along with the color change that occurs during each titration. Table 1. Potentiometric Titration of Unknown Acid
Trial 1 2 3
Volume of Volume of Mass of Moles of NaOH when NaOH to Unknown Color Change Color Change NaOH Reach Acid (g) Indicator Used Observed Observed (mL) Endpoint (mL) (mol) Bromothymol Blue 0.1158 Yellow-Blue 10.878 10.996 0.001096 0.1003 Phenolphtalein Clear-Pink 6.068 6.0157 0.0006016 Bromocresol YellowPurple Purple 6.327 6.357 0.0006357 0.103
3
The results of the identification of an unknown acid are shown in Table 2 below. The moles of the unknown acid by using the stoichiometry reaction to convert the moles of NaOH to moles of the unknown acid. The molar mass of the unknown acid was then calculated using Equation 2. Then the pKa of the unknown acid was calculated by finding the half-equivalence point from the titration graph for that particular trial, and the pH level that matched the halfequivalence point number. The average molar mass of the unknown acid was found to be 144.8 g/mol, the standard deviation was found to be 34.02 g/mol, and the percent relative standard deviation was found to be 23.49%. The 95% confidence limit for this data was found using Equation 3 and found to be ±38.50. The average of the pKa of the unknown acid was found to be 3.286, the standard deviation was 0.1280, and the percent relative standard deviation was 3.895%. The 95% confidence limit for this data was found using Equation 4 and found to be ±0.1448. Table 2. Identification of an Unknown Acid
Moles of Unknown Acid Trial 1: 0.001097 Trial 2: 0.0006016 Trial 3: 0.0006357 Average: Standard Deviation: % RSD 95% Confidence Limit
Molar Mass of Unknown Acid (g/mol) 105.6 166.7 162.0
pKa of Unknown Acid (from half-equivalence) 3.149 3.374 3.350
144.8 g/mol 34.02 g/mol 23.49% 38.50 g/mol
3.291 0.1236 3.895% 0.1448
The results for the titration curve of trial 2(Phenolphthalein) are shown in Table 3 below. This titration curve was created by taking the raw data collected from the MicroLab potentiometric titration, and then the student decimated the data. The decimated data was then processed through CuritPlot. By running this data through CuritPlot, a smooth titration curve was created, and other data was provided such as second derivative. The second derivative identified the endpoint volume for this specific titration trial. Through this titration curve graph the student was also able to find the pKa by finding the half-equivalence point, dividing that number by two, and then finding the pH that correlates with that number on the graph.
4
Table 3. Trial 2: Potentiometric Titration of an Unknown Acid
Discussion The purpose of this lab was to standardize AgNO3 and to find the mass and percent of Cl in an unknown sample. There was several errors that occurred in the students first titration, all of the data from this trial was erased and trial one was completed by someone else but the same method was used as the student for the second and third trial. The molar mass from trial one was vastly different from the student’s other data and the Grubbs Test was done, but the data did fit within the Grubbs test and could not be rejected. Based on the three titration trials, the results show that the average molar mass of the unknown acid was found to be 144.8 g/mol, standard deviation was 34.02 g/mol, and percent relative data was 23.49%. The 95% confidence limits for the concentration of the silver nitrate solution was found to be ±38.50 g/mol. The results also show that the average pKa for the unknown acid was 3.291. The standard deviation also calculated and was found to be 0.1236, and the percent relative standard deviation was found to be 3.895%. The 95% confidence limit for this data was found to be ±0.1448. The student then looked at the calculated averages for the molar mass and pKa of the unknown acid, these values were then compared to the four acids molar masses and pKa’s listed in Reference Table A, that the student was give in the in-lab procedure. By looking it was found that values of Mandelic Acid followed most closely with the values of the unknown acid that the student calculated. The values from this data set show that the measurements that the student made were close to one another, but they were not close to the value of first trial, completed by someone 5
else. It was interesting how the CurtiPlot was able to make such smooth titration curves compared to how the curve would look based solely off of the raw data points. It was also surprising, seeing that in the data that the student received for the first trial was vastly different from the other two trials in the molar mass, but they were all still similar in their pKa values. In most to all cases there is always some ways that errors can occur, the most common one that would apply to this experiment would be human error. The student could have recorded the wrong values or could have also recorded values that were past the endpoint for the titration. Other human errors include the possibility that the pH probe could have been calibrated incorrectly leading to incorrect pH readings or the amount of moles of NaOH could have been incorrectly calculated. Some systematic errors that could have occurred could be that the stopcock could be leaking, or the drop counter could not be miscounting, thus affecting the value for the volume needed to reach the endpoint. These could have occurred along with some other possible human and instrument errors; in this case it is likely that most of the possible errors came from the student.
Conclusion In this experiment, the overall objective was to understand the use of potentiometric titrations, and to understand the titration curve, pKa of an acid, to determine the molar mass of an unknown weak monoprotic acid, and to confirm the identity of the unknown monoprotic acid. This was accomplished by the potentiometric titration of the unknown acid with NaOH. The moles of the unknown were found by titrating the sample and using the volume needed to reach the endpoint and with the Molarity of NaOH, and then applying stoichiometry to find the moles of the unknown acid. The moles and mass of the unknown acid was then used to find the molar mass of the unknown. The results showed the average molar mass of the unknown acid to be 144.8 g/mol, the standard deviation of the unknown was then found to be 34.02 g/mol. The precent relative standard deviation and confidence level at 95% were also found to be 23.49% and ±38.50 g/mol. Then the pKa of each of the three titrations was calculated by using the halfequivalence point and pH in the titration curve. The average pKa of the unknown acid was found to be 3.291, the standard deviation was found to be 0.1236, and the percent relative standard deviation was found to be 3.895%. The 95% confidence limit for this data was found ±0.1448. Then the average molar mass and pKa was compared to the possible unknown acids from Table
6
A. The acid that is closest to the values found with the unknown was Mandelic Acid with a molar mass of 152.15 and a pKa of 3.41. Therefore, the identification of the Unknown acid is Mandelic Acid.
Sample Calculations a. Moles of NaOH
= ( ) × ( )
.
= . × . = . = ±. b. Molar Mass of Unknown
# = #
.
. = . / . = . / c. Confidence Level at 95% of Molar Mass of Unknown Acid
� � ±= √ . ±
.
. × . √
d. 95% Confidence Interval of pKa of Unknown Acid
� � ±= √ . ±
.
. × . √
= ±.
7
References [1] Harris, D. C. Quantitative Chemical Analysis, 8th ed.; W.H. Freeman: New York, 2010; p 56. [2] Soldatov, V. S. "Potentiometric titration of ion exchangers." Reactive and Functional Polymers 38.2-3 (1998): 73-112. [3] Libretexts. (2020, August 11). 8.3: PKa Values. Retrieved October 28, 2020, from https://chem.libretexts.org/Courses/Purdue/Purdue:_Chem_26505:_Organic_Chemistry_I_(Lipto n)/Chapter_8._Acid-Base_Reactions/8.3:_pKa_Values
8...
Similar Free PDFs
Popular Institutions
- Tinajero National High School - Annex
- Politeknik Caltex Riau
- Yokohama City University
- SGT University
- University of Al-Qadisiyah
- Divine Word College of Vigan
- Techniek College Rotterdam
- Universidade de Santiago
- Universiti Teknologi MARA Cawangan Johor Kampus Pasir Gudang
- Poltekkes Kemenkes Yogyakarta
- Baguio City National High School
- Colegio san marcos
- preparatoria uno
- Centro de Bachillerato Tecnológico Industrial y de Servicios No. 107
- Dalian Maritime University
- Quang Trung Secondary School
- Colegio Tecnológico en Informática
- Corporación Regional de Educación Superior
- Grupo CEDVA
- Dar Al Uloom University
- Centro de Estudios Preuniversitarios de la Universidad Nacional de Ingeniería
- 上智大学
- Aakash International School, Nuna Majara
- San Felipe Neri Catholic School
- Kang Chiao International School - New Taipei City
- Misamis Occidental National High School
- Institución Educativa Escuela Normal Juan Ladrilleros
- Kolehiyo ng Pantukan
- Batanes State College
- Instituto Continental
- Sekolah Menengah Kejuruan Kesehatan Kaltara (Tarakan)
- Colegio de La Inmaculada Concepcion - Cebu