Activity 6 - Tree Thinking PDF
| Title | Activity 6 - Tree Thinking |
|---|---|
| Author | shwikar Abdelrahman |
| Course | Human Genetics |
| Institution | University of Maryland Baltimore County |
| Pages | 7 |
| File Size | 343.7 KB |
| File Type | |
| Total Downloads | 63 |
| Total Views | 171 |
Summary
Download Activity 6 - Tree Thinking PDF
Description
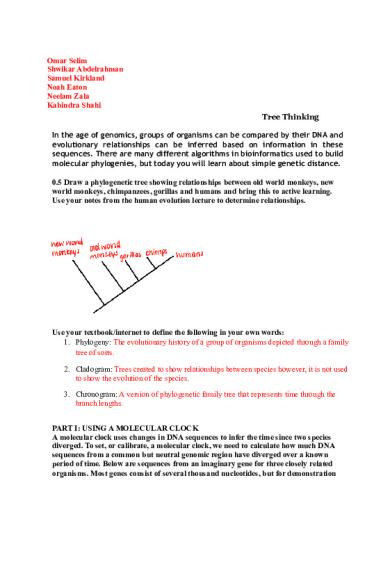
Omar Selim Shwikar Abdelrahman Samuel Kirkland Noah Eaton Neelam Zala Kabindra Shahi Tree Thinking In the age of genomics, groups of organisms can be compared by their DNA and evolutionary relationships can be inferred based on information in these sequences. There are many different algorithms in bioinformatics used to build molecular phylogenies, but today you will learn about simple genetic distance. 0.5 Draw a phylogenetic tree showing relationships between old world monkeys, new world monkeys, chimpanzees, gorillas and humans and bring this to active learning. Use your notes from the human evolution lecture to determine relationships.
Use your textbook/internet to define the following in your own words: 1. Phylogeny: The evolutionary history of a group of organisms depicted through a family tree of sorts. 2. Cladogram: Trees created to show relationships between species however, it is not used to show the evolution of the species. 3. Chronogram: A version of phylogenetic family tree that represents time through the branch lengths. PART I: USING A MOLECULAR CLOCK A molecular clock uses changes in DNA sequences to infer the time since two species diverged. To set, or calibrate, a molecular clock, we need to calculate how much DNA sequences from a common but neutral genomic region have diverged over a known period of time. Below are sequences from an imaginary gene for three closely related organisms. Most genes consist of several thousand nucleotides, but for demonstration
purposes, we are only going to use some of the bases. Calculate the percentage sequence divergence between all three sequences. First examine the sequences in question and count the differences between them. Divide the difference by the total number of nucleotides, and then multiply by 100 to convert to a percent. DNA Sequence of Cyt-b (Mitochondrial DNA) Human ACTAGCAACGGATACCATAGGTATATCTAGGCTACATTGT Chimp ACTAGAAACGGTTACCACAGGTATATCAAGGCAACATTGT Baboon ACTGGATGCGGATGCCTCAGATTTATCAAGGCTACAATGT
Human Chimp Baboon
DNA Sequence of Cyt-b (Mitochondrial DNA) continued TAGCTTACCGCTAGTACTGGTGACTCTAGAATGCCTAGTC TACCTTACCGCTAGTACTGGTGACTCTCGAATGCCTTGTC TACCATAGCGATACTTGTGGTGACTGTCGATTGCCTAGTG
4. Human and Chimp: 10% •
8 differences/80 nucleotides*100 = 10%
5. Chimp and Baboon: 26.25% • 21 differences/80 nucleotides*100 = 26.25%
6. Baboon and Human: 27.5% • 22 differences/80 nucleotides*100 = 27.5% The major assumption of a molecular clock is that sequence divergence increases over time in a linear manner as species diverge. This means that species that diverged 4 million years ago should have twice the sequence divergence as species that diverged 2 million years ago. If one knows when two species diverged (by using the dates of fossils, geological events, etc.), one can use this information to calibrate a molecular clock for that particular region of the genome. That clock can then be applied to other species and used to determine when other species diverged. Use the equation D=μt where D=% sequence divergence, μ=mutation rate, and t=time since divergence for the next 4 questions. 7. First calculate the mutation rate for each species pair below. What is the average mutation rate, μ (% divergence per million years), for this particular gene? Species-Pair Sequence Time Since μ Divergence Divergence (yrs) 1.78 Human-Chimp 9.8% 5.5 million 1.71 Human12.0% 7.0 million Gorilla
Chimp-Gorilla
12.8%
7.0 million
1.83
Average μ = 1.77
8. If the sequence divergence between chimpanzees and another great ape group called bonobos at this gene is 4.2%, calculate the estimated time since divergence between chimps and bonobos. 4.2/1.8= 2.4 million years ago 9. A strict perfect molecular clock predicts that the human-gorilla percent sequence divergence should be exactly the same as the chimp-gorilla percent sequence divergence. [Think about why that should be; drawing a tree of the three species should help.] However, there is a difference in the above table. Propose a potential explanation for why the % sequence divergence differs between human-gorilla and chimp- gorilla even though time since divergence is the same. The human gorilla divergence sequence is exactly same as that of chimp-gorilla sequence divergence.There is some difference in % sequence divergence as from protein analysis we can say that point mutation generate some variations .The analysis shows some results which depict that the gorilla is the closest ancestor of both humans and chimpanzees, further analysis showed that chimpanzees to be closer relative of gorilla than humans. 10. APPLYING WHAT YOU KNOW: The sequence divergence between modern H. sapiens and Neanderthals (based on DNA obtained from fossils) is 1.2% at this gene. How long ago did modern H. sapien and Neanderthal share a common ancestor? 1.2/1.8 = 0.67 million years ago 11. Construct an UNSCALED TREE (cladogram) showing the relationship between humans, chimps, bonobos, and gorillas using this sequence data (draw an arrow to indicate the TIME axis). One person from each table should volunteer and draw this cladogram on the board for the whole table. (Compare your nodes [there is only one correct tree] and your tree drawing style [many possibilities]).
Bon obos Goril las Hum ans
Chi mps
12. Now construct a SCALED TREE (chronogram) showing the relationship between humans, chimps, bonobos, and gorillas using this sequence data. How is it different from your cladogram?
It’s different from the cladogram because the chronogram is scaled based on time and the branch lengths differ based on the time.
PARSIMONY: the most likely explanation implies the least amount of change; the simplest explanation is preferred. 13. Do you think the common ancestor to humans, chimps, bonobos and gorillas walked full-time upright on two feet (biped)? Use parsimony to explain. Parsimony shows that the common ancestor of humans and all the apes lived in Europe and bipedalism originated in that ancestor. 14. Do you think the common ancestor to humans, chimps, bonobos and gorillas had body covered with hair? Use parsimony to explain. I think the common ancestor didn't have a body covered with hairs as some mutation i.e. hairless mutation might have occured in the common ancestor which diverged humans and other apes. The great ancestor of humans and apes might have had the hairs. PART II. BASED ON PROTEIN SEQUENCE DATA 15. The amino acid sequence for cyt-c gene is given for several species at the end of this handout. How many differences are there: ● Between humans and rhesus monkeys? 1 ● Between humans and chickens/turkeys? 7 ● Between humans and ducks? 6 ● ● ● ● ● ●
Between humans and Neurospora? 29 Between rhesus monkeys and chickens/turkeys? 6 Between rhesus monkeys and ducks? 5 Between rhesus monkeys and Neurospora? 28 Between ducks and chickens/turkeys? 1 Between Neurospora and baker’s yeast? 26
16. Draw a CLADOGRAM showing how human, rhesus monkey, duck, chicken/turkey, and Neurospora are
related to each other.
17. Explain how the chicken-turkey relationship is possible given that they are classified as different species. Why can two species have no divergence on our tree? Chicken and turkey are in the same tree because their amino acid sequence for cytochrome C gene/protein is the same, considering only that one gene. However in other proteins, there could be differences so they are different species. 18. Neurospora and Baker’s Yeast are both fungi and humans and monkeys are both primates. Assuming a mutation rate of 0.068 % divergence/million years, how long ago did Neurospora and Baker’s Yeast diverge according to the data for the cyt-c gene? What can you say about the divergence time between Neurospora and Baker’s Yeast compared to humans and rhesus monkeys? 26 nucleotide difference / 60 total nucleotides = 0.433 43.3% / 0.068% = 636.76 million years 1 nucleotide difference / 60 total nucleotides = 0.016 1.6% / 0.068% = 23.53 million years The divergence between Baker’s Yeast and Neurospora compared to humans and rhesus monkeys is much longer in terms of time because there are more differences between Neurospora and Baker’s Yeast than humans and rhesus monkeys according to data for the cyt-c gene.
AMINO ACID SEQUENCES IN CYTOCHROME-C PROTEINS FROM DIFFERENT SPECIES Amino Acid Number
1
2
3
4
5
6
7
8
9
0
1
2
3
4
5
6
7
8
9
0
1
2
3
4
5
6
7
8
9
0
Human
G
D
V
E
K
G
K
K
I
F
I
M
K
C
S
Q
C
H
T
V
E
K
G
G
K
H
K
T
G
P
Rhesus monkey
G
D
V
E
K
G
K
K
I
F
I
M
K
C
S
Q
C
H
T
V
E
K
G
G
K
H
K
T
G
P
Dog
G
D
V
E
K
G
K
K
I
F
V
Q
K
C
A
Q
C
H
T
V
E
K
G
G
K
H
K
T
G
P
Kangaroo
G
D
V
E
K
G
K
K
I
F
I
M
K
C
A
Q
C
H
T
V
E
K
G
G
K
H
K
T
G
P
Chicken, Turkey
G
D
I
E
K
G
K
K
I
F
V
Q
K
C
S
Q
C
H
T
V
E
K
G
G
K
H
K
T
G
P
Penguin
G
D
I
E
K
G
K
K
I
F
V
Q
K
C
S
O
C
H
T
V
E
K
G
G
K
H
K
T
G
P
Pekin duck
G
D
V
E
K
G
K
K
I
F
V
Q
K
C
S
Q
C
H
T
V
E
K
G
G
K
H
K
T
G
P
Bullfrog
G
D
V
E
K
G
K
K
I
F
V
Q
K
C
A
Q
C
H
T
C
E
K
G
G
K
H
K
V
G
P
Silkworm moth
G
N
A
E
N
G
K
K
I
F
V
Q
R
C
A
Q
C
H
T
V
E
A
G
G
K
H
K
V
G
P
Fungus 1 (Neurospora)
G
S
S
K
K
G
A
N
L
F
K
T
R
C
A
E
C
H
G
E
G
G
N
L
T
Q
K
I
G
P
Fungus 2 (baker’s yeast)
G
S
A
K
K
G
A
T
L
F
K
T
R
C
E
L
C
H
T
V
E
K
G
G
P
H
K
V
G
P
Amino Acid Number (continued)
1
2
3
4
5
6
7
8
9
0
1
2
3
4
5
6
7
8
9
0
1
2
3
4
5
6
7
8
9
0
Human
N
L
H
G
L
F
G
R
K
T
G
Q
A
P
G
Y
S
Y
T
A
A
N
K
N
K
G
I
I
W
G
Rhesus monkey
N
L
H
G
L
F
G
R
K
T
G
Q
A
P
G
Y
S
Y
T
A
A
N
K
N
K
G
I
T
W
G
Dog
N
L
H
G
L
F
G
R
K
T
G
Q
A
P
G
F
S
Y
T
D
A
N
K
N
K
G
I
T
W
G
Kangaroo
N
L
N
G
I
F
G
R
K
T
G
Q
A
P
G
F
T
Y
T
D
A
N
K
N
K
G
I
I
W
G
Chicken, Turkey
N
L
H
G
L
F
G
R
K
T
G
Q
A
E
G
F
S
Y
T
D
A
N
K
N
K
G
I
T
W
G
Penguin
N
L
H
G
I
F
G
R
K
T
G
Q
A
E
G
F
S
Y
T
D
A
N
K
N
K
G
I
T
W
G
Pekin duck
N
L
H
G
L
F
G
R
K
T
G
Q
A
E
G
F
S
Y
T
D
A
N
K
N
K
G
I
T
W
G
Bullfrog
N
L
Y
G
L
I
G
R
K
T
G
Q
A
A
G
F
S
Y
T
D
A
N
K
N
K
G
I
T
W
G
Silkworm moth
N
L
H
G
F
Y
G
R
K
T
G
Q
A
P
G
F
S
Y
S
N
A
N
K
A
K
G
I
T
W
G
Fungus 1 (Neurospora)
A
L
H
G
L
F
G
R
K
T
G
S
V
D
G
Y
A
Y
T
D
A
N
K
Q
K
G
I
T
W
D
Fungus 2 (baker’s yeast)
N
L
H
G
I
F
G
R
H
S
G
Q
A
Q
G
Y
S
Y
T
D
A
N
I
K
K
N
V
L
W
D
AMINO ACID SYMBOLS A = Alanine C = Cysteine D = Aspartic acid E = Glutamic acid F = Phenylalanine G = Glycine
H I K L M N P
= = = = = = =
Histidine Isoleucine Lysine Leucine Methionine Asparagine Proline
Q R S T V W Y
= = = = = = =
Glutamine Arginine Serine Threonine Valine Tryptophan Tyrosine...
Similar Free PDFs

Activity 6 - Tree Thinking
- 7 Pages

Critical Thinking Assignment 6
- 2 Pages

Critical Thinking Assignment 6-1
- 1 Pages

Critical Thinking Assignment 6-3
- 3 Pages

Critical Thinking Assignment 6-2
- 2 Pages

Critical Thinking Assignment 6-4
- 2 Pages

Fraud Tree
- 10 Pages

Income Taxation Activity 6
- 3 Pages
Popular Institutions
- Tinajero National High School - Annex
- Politeknik Caltex Riau
- Yokohama City University
- SGT University
- University of Al-Qadisiyah
- Divine Word College of Vigan
- Techniek College Rotterdam
- Universidade de Santiago
- Universiti Teknologi MARA Cawangan Johor Kampus Pasir Gudang
- Poltekkes Kemenkes Yogyakarta
- Baguio City National High School
- Colegio san marcos
- preparatoria uno
- Centro de Bachillerato Tecnológico Industrial y de Servicios No. 107
- Dalian Maritime University
- Quang Trung Secondary School
- Colegio Tecnológico en Informática
- Corporación Regional de Educación Superior
- Grupo CEDVA
- Dar Al Uloom University
- Centro de Estudios Preuniversitarios de la Universidad Nacional de Ingeniería
- 上智大学
- Aakash International School, Nuna Majara
- San Felipe Neri Catholic School
- Kang Chiao International School - New Taipei City
- Misamis Occidental National High School
- Institución Educativa Escuela Normal Juan Ladrilleros
- Kolehiyo ng Pantukan
- Batanes State College
- Instituto Continental
- Sekolah Menengah Kejuruan Kesehatan Kaltara (Tarakan)
- Colegio de La Inmaculada Concepcion - Cebu







