Introduction to Computing, Homework 7, Questions and Answers PDF

| Title | Introduction to Computing, Homework 7, Questions and Answers |
|---|---|
| Course | Introduction to Computing |
| Institution | Johns Hopkins University |
| Pages | 10 |
| File Size | 295.5 KB |
| File Type | |
| Total Downloads | 16 |
| Total Views | 119 |
Summary
This document contains the 7th homework for the Introduction to Computing course from Fall 2016. This document is highly useful in preparation for the exams in this course....
Description
Answers in yellow Comments in green. Established and discussed during grading session
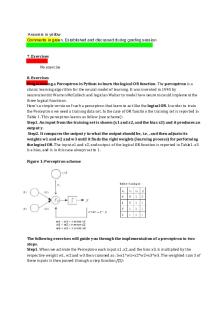
7.#Exercises No#exercise 8.#Exercises Programming#a#Perceptron#in#Python#to#learn#the#logical#OR#function.#The#perceptron#is#a# classic#learning#algorithm#for#the#neural#model#of#learning.#It#was#invented#in#1943#by# neuroscientist#Warren#McCulloch#and#logician#Walter#to#model#how#neurons#could#implement#the# three#logical#functions. Here's#a#simple#version#of#such#a#perceptron#that#learn#to#act#like#the#logical#OR.#In#order#to#train# the#Perceptron#we#need#a#training#data#set.#In#the#case#of#OR#function#the#training#set#is#reported#in# Table#1.#This#perceptron#learns#as#follow#(see#scheme): Step1.#An#input#from#the#training#set#is#shown#(x1#and#x2,#and#the#bias#x3)#and#it#produces#an# output#y. #Step2.#It#compares#the#output#y#to#what#the#output#should#be,#i.e.#,#and#then#adjusts#its# weights#w1#and#w2#and#w3#until#it#finds#the#right#weights#(learning#process)#for#performing# the#logical#OR.#The#input#x1#and#x2,#and#output##of#the#logical#OR#function#is#reported#in#Table1.#x3# is#a#bias,#and#is#in#this#case#always#set#to#1. # Figure#1:#Perceptron#scheme
The#following#exercises#will#guide#you#through#the#implementation#of#a#perceptron#in#two# steps. Step1.#When#we#activate#the#Perceptron#each#input#x1,#x2,#and#the#bias#x3,#is#multiplied#by#the# respective#weight#w1,#w2#and#w3#then#summed#as:#S=x1*w1+x2*w2+x3*w3.#The#weighted#sum#S#of# these#inputs#is#then#passed#through#a#step#function#f(S): ############ ##################################################
The#output#of#this#function#is#the#output#of#the#Perceptron.,#which#has#only#two#possible#values#:#0#and#1.#The# bias,#x3,#is#a#"dummy"#input#that#is#needed#to#move#the#threshold#as#needed#by#the#step#function.#Its#value#is# always#x3=1,#so#that#its#influence#on#the#result#can#be#controlled#by#its#weight#w3.
1.#(10)#Write#a#python#module#(perc_func.py)##that#contains#the#following#functions: ### a.#(2)#Make#a#function#called#weight_sum#that#takes#in#6#arguments:#x1,8x2,8x3,8w1,8w2#and#w3# and#returns#the#weighted#sum#S,#i.e.#S=x1*w1+x2*w2+x3*w3 ### b.#(3)#Make#a#function#called#step_func#that#takes#in#one#argument,#the#weighted#sum##S,#and# returns#1#if#the#sum#is#greater#than#or#equal#to#0,#otherwise#have#it#return#0. ### c.#(3)#Make#a#function#called#perc_output#that#takes#x1,8x2,8x3,8w1,8w2#and#w3#and#returns#the# output#y#by#using#functions#step_func#and#weight_sum. ## e.#(2)#Test#your#module#with#input#x1=0,#x2=1,#x3=1,#w1=0.2,#w2=0.4#and#w3=0.2#and#print# the#output#of#the#function#perc_output.#Make#sure#you#do#your#testing#after#: if#__name__#==#"__main__": #def#weight_sum(x1,x2,x3,w1,w2,w3): ####return#x1*w1+x2*w2+x3*w3 def#step_func(S): ####if#S#>=0.0: #######return#1 ####else: ########return#0
If#they#have#S>=#1#we#should#accept#because#there#was#a#typo#in#the#original#version#of#the#HW. Check#this#if#the#output#is#0. -3#if#they#print#and#do#not#return# def#perc_output(x1,x2,x3,w1,w2,w3): ####S1=weight_sum(x1,x2,x3,w1,w2,w3) ####return#step_func(S1) if#__name__#==#"__main__": ####print(perc_output(0,1,1,0.2,0.4,0.2)) Output is 1 -0 if took values as input from command line -2 for only having a print statement under the if and running function outside of if -0 for returning 1 if the sum is greater than or equal to 1 Same#as#last#time#-#1#point#per#syntax#error#-#up#to#half#points,#round#down#(e.g.,#if#it’s#3#point# questions,#they#can#lose#2#points)#Up#to#a#maximum#of#10#points#deducted#for#repeated#mistakes! -2#for#return#step_func(weight_sum)#with#no#arguments#to#weight_sum# -1#for#using#sys.argv#but#not#importing#sys# -1#for#converting#S#to#int# -1#does#not#print#output#in#if#main#
Step2. Compare#the#output#y#to#the#expected#value.#This#will#produce#an#error,#i.e.#8.#To#find#the#weights#that# give#rise#to#the#logical#OR,#we#have#to#adjust#the#weights#based#on#the#error#and#the#learning#rate#r.#This#is# repeated#until#(in#this#case)#a#maximum#number#of#iterations#n#is#reached.
2.#(25)#Make#a#script#called#perceptron.py#and#write#the#following: ######### a.#(1)##Import#module#perc_func.py8and#the#random#module import perc_func import random #print(perc_func.output(1,0,0.6,0.6)) #########
b.#(2)#Generate#three#different#real#random#numbers#w1,8w2#and#w3#from#0#to#1.
w1=random.uniform(0,1) w2=random.uniform(0,1) w3=random.uniform(0,1) OR random.random() -1 for using random.randint(0,1) ########## c.#(1)#Define#variable#r=0.2 r=0.2 ########## d.##(3)#Make#a#tuple#of#tuples:#T8=((0,0,1,#0),(1,0,1,1),(0,1,1,1),(1,1,1,1))#which#is#the#training# set#reported#in#Table1.#Note#that#the#4#values#in#each#of#the#4#tuples#describe#values#(x1,8x2,8x3,# ybar).#The#bias#x3#is#always#set#to#1. #print(w1,w2) #T=((x1,x2,x3,y),...) T=((0,0,1,0),(1,0,1,1),(0,1,1,1),(1,1,1,1)) ######### e.#(2)#Make#a#for#loop#of#n=200#and#for#each#cycle: ########## f.#(3)#Use#a#method#from#the#random#module#to#pick#one#of#the#4#(nested)#tuples#from#the# big#tuple#T.#Note#that#your#random#pick#will#be#a#tuple,#since#T#is#a#tuple#of#tuples.#You#can#store#the# randomly#chosen#tuple#in#a#variable#called#my_t. ###############g.#(2)#Store#the#4#values#from#my_t#in#variables#x1,8x2,8x3#and#ybar. ############# h.#(2)#Use#the#perc_output#function#with#x1,8x2,8x3,8w1,8w2#and#w3#and#store#the#results#in#a# variable#called#y. -2#for#perc_output#and#not#putting#perc_func.perc_output(...)# Unless#they#did#from#perc_func#import#perc_output ################i.##(1)#Calculate#the#difference#between#the#expected#value#ybar#and#the#output#of#the# perceptron#y#and#store#the#result#in#a#variable#called#perc_8error ############## j.##(2)#Adjust#weights#as#:
##############################w1=w1+r*perc_error*x1 888888888888888888888888888888w2=w2+r*perc_error*x2 888888888888888888888888888888w3=w3+r*perc_error*x3 ########################### for n in range(0,200): my_t=random.choice(T) x1=my_t[0] x2=my_t[1] x3=my_t[2] ybar=my_t[3] y=perc_func.perc_output(x1,x2,x3,w1,w2,w3) perc_error=ybar-y w1=w1+r*perc_error*x1 w2=w2+r*perc_error*x2 w3=w3+r*perc_error*x3 print(error,y,x1,x2,x3,w1,w2,w3) -0 for omitting this line -1 for range(201) -1 for using random.random(0,3) instead of randint
# k.#(1)#Outside#the#loop#print#out#the#final#(learned)#weights#w1,#w2#and#w3.#Print#a#formatted#output# with#3#decimal#digits. print(w1,w2,w3) -1 if output not formatted to 3 decimal places -- only worth 1pt l.#(5)#To#test#the#perceptron,#use#the#final#learned#w1,w2#and#w3#on#each#of#the#training#set#values# (x1,x2,x3)#from#the#tuple#T,#by#using#the#perc_output#function.#For#each#training#set,#print#x1,8x2#and# the#output#of#the#perc_output8function#to#confirm#whether#such#weights#will#reproduce#the#OR# function#(Table#1).#Use#for#loop#over#the#tuple#T.#Remember#that#T#is#a#nested#tuple. You#should#get#an#output#like#this: 0#0#0 1#0#1 0#1#1 1#1#1 It#worked!#You#have#just#implemented#your#first#rudimental#learning#algorithm!! for t in T: x1=t[0] x2=t[1] x3=t[2]
print(x1,x2,perc_func.output(x1,x2,x3,w1,w2,w3)) -2 If they do not have last for loop (we asked explicitly for for loop) -0 for getting inconsistent results because of the HW typo error in part 1b -1 syntax errors (indentation, :) -1 for T[t][0] -1 for missing perc_func. -1 used same x3 every time instead of t[2] -1 missing t in loop initialization -1 for range(1,4) (should be 0,4) #
9.#Exercises 1. (60#points#)#Write#a#program#(amminoacid_stat.py)#that#analyzes#protein#data#bank#file#for# sequence#information.#We#are#providing#you#with#a#protein#data#file#cry_atoms.pdb.8We#worked# with#similar#files#in#the#first#part#of#this#course.#The#1st##column#contains#a#keyword#ATOM,#2nd# column#atom#numbers,#3rd#column#atom#types,#4th#column#amino#acid#names,#and#5th#column#chain# name.#The#protein#provided#has#only#two#chains,#A#and#B,#so#5th#column#can#take#up#only#two# values,#A#or#B. (Always#view#your#data#file#before#starting#manipulating#it.#You#can#use#an#appropriate#bash# command.) The#program#should#do#the#following: a. #(8)#Read#in#the#provided#protein#sequence#data#file#(cry_atoms.pdb).##Read#in#all#of#the#lines# from#that#file,#and#store#the#lines#in#a#list#called#atomlist f1=open('cry_atoms.pdb','r') atomlist=f1.readlines() f1.close() -0#For#not#opening#file#in#‘read’#mode#--#still#works -0#if#they#do#not#close#file.#It#will#still#work.# -1#per#syntax#error#-#same#as#before
b. (2)#Print#the#first#item#in8atomlist.8The#output#should#be: 'ATOM
1 N ALA A 2
-31.589 -7.997 48.685 1.00 59.63
N \n'
atomlist[0] -0#It#is#OK#if#they#do#print(atomlist[0])# # c. (8)#Generate#a#list#(amm_list)#that#contains#only#lines#that#have#“CA”#in#the#3rd##column. amm_list#=#[] for#i#in#atomlist: ####if#i.split()[2]#==#"CA":
########amm_list.append(i) 2#points#for#initiating##empty#list 2#for#for#loop 3#for#append 3#for#correct#conditional -0.1#if#they#don’t#query#3rd#column,#e.g,##if#‘CA’#in#line…#Not#robust# d. (2)#Use#print#function#3#times#in#a#row#to#print#the#first#3#items#in#amm_list.#The#output#should# be: 'ATOM 'ATOM 'ATOM
2 CA ALA A 2 -31.278 -6.672 48.156 1.00 65.73 7 CA THR A 3 -29.720 -7.309 44.783 1.00 62.19 14 CA ARG A 4 -28.711 -3.785 43.697 1.00 53.94
C \n' C \n' C \n'
#
amm_list[0] amm_list[1] amm_list[2] -0#It#is#OK#if#they#do#print(amm_list[0])# -0#for#amm_list[:1],#amm_list[1:2],#etc# e. (2)#Initialize#an#empty#dictionary##d.8This#dictionary#should#have#two#keys#that#correspond#to# strings#“A”#and#“B”.#For#each#of#the#keys#initialize#an#empty#list#as#a#value. d={}# d['A']=[] d['B']=[] 2#points#for#the#above It’s#OK#if#they#do#it#later. -1#for#d[“A”]={}#instead#of#[]# f. (8)##Note#that#keys#“A”#and#“B”#correspond#to#chain#names#that#are#found#in#the#5 th#column.#The# list#that#correspond#to#dictionary#key#“A”#will#contain#a#sequence#of#amino#acid#names#(found#in#the# 4th#column)#of#chain#A,#and#the#list#that#correspond#to#dictionary#key#“B”#will#contain#a#sequence#of# amino#acid#names#(found#in#the#4th#column)#of#chain#B.#You#should#generate#these#lists.#As#an# example,#the#first#3#amino#acids#in#the#list#corresponding#to#key#“A”#should#be:#“ALA”,#“THR”,#“ARG”. for#line#in#range(0,len(amm_list)): ####if#amm_list[line].split()[4]=='A': #######d['A'].append(amm_list[line].split()[3]) ####else: #######d['B'].append(amm_list[line].split()[3]) OR for#i#in#amm_list:
####if#i.split()[4]#==#"A": ########d['A'].append(i.split()[3]) ####elif#i.split()[4]#==#"B": ########d['B'].append(i.split()[3])
2#points#for#loop 3#points#correct#append 3#points#correct#conditional -1#for#i,split()# -0#for#initializing#empty#lists,#appending#those,#and#setting#d[‘A’]#=#listA# -2#missing#the#split#so#the#list#is#wrong
g. (8)#Count#how#many#times#the#keyword#“GLU”#appears#in#dictionary#d.#Store#the#result#in# another#dictionary#called#d_count,#that#contains#the#same#2#keys,#“A”#and#“B”,#and#as#corresponding# values#another#dictionary#(one#for#each#chain).#This#new#dictionary#should#contain#one#key,#“GLU”# and#as#value#it#should#contain#the#corresponding#count#for#each#chain.#Do#not#forget#to#initialize#any# new#dictionaries. countA#=#0 for#i#in#d['A']: ####if#i#==#"GLU": ########countA#=#countA#+#1 countB#=#0 for#i#in#d['B']: ####if#i#==#"GLU": ########countB#=#countB#+#1 d_count#=#{}## d_count['B']#=#{} d_count['A']#=#{}
d_count['B']['GLU']#=#countB d_count['A']['GLU']#=#countA OR d_count={} d_count["A"]={} d_count["B"]={} d_count["A"]["GLU"]=d["A"].count('GLU')
d_count["B"]["GLU"]=d["B"].count('GLU') -NO#POINTS#Awarded#for#hard-coding#the#dictionary
h. (2)#Repeat#the#same#for#keyword##“LYS”.#If#you#do#print(d_count)#the#answer#should#be: {'B': {'LYS': 15, 'GLU': 36}, 'A': {'LYS': 15, 'GLU': 36}}
Note#that#the#order#of#appearance#of#A#and#B#as#well#as#LYS#and#GLU#in#the#dictionary#does#not# matter. If#you#have#trouble#completing#this#part,#simply#generate#d_count#by#typing#in#the#above#dictionary# for#a#loss#of#all#points#on#parts#g.#and#h.#You#will#still#be#able#to#continue#with#parts#i,#j,#k. countA#=#0 for#i#in#d['A']: ####if#i#==#"LYS": ########countA#=#countA#+#1 countB#=#0 for#i#in#d['B']: ####if#i#==#"LYS": ########countB#=#countB#+#1 d_count['B']['LYS']#=#countB d_count['A']['LYS']#=#countA This#will#be#tough#to#grade#as#there#are#many#ways#to#do#this.# Glu#and#Lys#should#add#up#to#a#total#of#10#points.#Here#is#a#guess: 1#points#for#correct#initialization#of#dictionaries.# 1#points#to#initialize#the#count#variable 2#points#correct#if#statements 2#points#to#increase#the#count 2#points#for#correctly#assigning#values#to#first#dictionary 2#points#for#correctly#assigning#values#to#dictionary#inside#dictionary -NO#POINTS#Awarded#for#hardcoding#the#dictionary -1#for#not#adding#GLU#key#and#count#value#to#the#dictionary#but#did#wit#LYS i. (2)#Check#if#the#two#dictionaries#within#d_count#are#the#same. d_count['A']#==#d_count[‘B’] 2#points# -0#if#the#student#correctly#uses#cmp#function,#i.e.#cmp(d_count[‘A’],d_count[‘B’])#### j.
(9)#Use#looping#through#dictionary#d_count#to#print#the#following#statement: For chain B, and residue LYS, the count is 15. For chain B, and residue GLU, the count is 36. For chain A, and residue LYS, the count is 15.
For chain A, and residue GLU, the count is 36.
Note#that#the#order#of#appearance#of#A#and#B#as#well#as#LYS#and#GLU#does#not#matter.#You#should# not#hardcode#A,#B,#LYS,#GLU#or#any#of#the#numbers. for#i#in#d_count: ####for#j#in#d_count[i]: ########print('For#chain#%s,#and#residue#%s,#the#count#is#%d.'#%(i,j,d_count[i][j])) 4#points#for#first#for#loop They#do#not#need#to#have#second#loop.#This#will#be#an#easy#one -1#for#d_count(i)# -0##if#doesn’t#print#only#because#dictionary#was#not#properly#formed#in#part#g#and#h# k. (9)#Write#the#same#statements#in#a#file#called#cry_amm_stat.dat#with#the#write#method. f3=open('cry_amm_stat.dat','w') for#i#in#d_count: ####for#j#in#d_count[i]: ########f3.write('For#chain#%s,#and#residue#%s,#the#count#is#%d.\n'#%(i,j,d_count[i][j])) f3.close() 2#points#correct#open 3#points#correct#close 4#points#correct#everything#else -0#for#hardcoding -0#########For#used#readlines#to#copy##the#aminoacid_stat.py##script#into#cry_mm_stat.dat…# -0#????#for#missing#\n# l. (5)#Extra#credit#question. Create#dictionary#d_count_all#which#is#similar#to#d_count,#only#it#contains#information#for#all#amino# acids#(not#just#GLU#and#LYS).#Note#that#there#are#20#amino#acids,#and#they#are#all#contained#at#least# once#in#each#of#the#lists#in#dictionary#d.#You#should#not#do#this#by#manually#adding#each#amino#acid# as#we#did#for#the#case#of#GLU#and#LYS,#but#should#use#multiple#nested#loops.
##create#a#list#of#amino#acids my_aa#=#[] for#i#in#d['A']: ####while#my_aa.count(i)#==#0: ########my_aa.append(i) ##initialize#empty#dictionaries d_count#=#{} for#c#in#'AB': ####d_count[c]#=#{}
for#aa#in#my_aa: ####for#c#in#'AB': ########count#=#0 ########for#i#in#d[c]: ############if#i#==#aa: ################count#=#count#+#1 ########d_count[c][aa]#=#count print(d_count) ####### 1#pt#for#attempt#at#extra#credit#that#does#not#run No#credit#for#manually#adding#every#amino#acid -2#for#hardcoding#list#of#amino#acids Correct:#5,#attempt:#1,#incorrect:#0...
Similar Free PDFs

Introduction to computing final exam
- 19 Pages

Homework 4 - Questions and answers
- 10 Pages

Introduction to Cakes Homework
- 42 Pages

Chapter 7 Homework Answers
- 37 Pages

7 2020, questions and answers
- 13 Pages

Chapter 7 Questions and Answers
- 48 Pages

Chapter 7 Homework Questions
- 5 Pages

Homework for Introduction to CVT
- 2 Pages
Popular Institutions
- Tinajero National High School - Annex
- Politeknik Caltex Riau
- Yokohama City University
- SGT University
- University of Al-Qadisiyah
- Divine Word College of Vigan
- Techniek College Rotterdam
- Universidade de Santiago
- Universiti Teknologi MARA Cawangan Johor Kampus Pasir Gudang
- Poltekkes Kemenkes Yogyakarta
- Baguio City National High School
- Colegio san marcos
- preparatoria uno
- Centro de Bachillerato Tecnológico Industrial y de Servicios No. 107
- Dalian Maritime University
- Quang Trung Secondary School
- Colegio Tecnológico en Informática
- Corporación Regional de Educación Superior
- Grupo CEDVA
- Dar Al Uloom University
- Centro de Estudios Preuniversitarios de la Universidad Nacional de Ingeniería
- 上智大学
- Aakash International School, Nuna Majara
- San Felipe Neri Catholic School
- Kang Chiao International School - New Taipei City
- Misamis Occidental National High School
- Institución Educativa Escuela Normal Juan Ladrilleros
- Kolehiyo ng Pantukan
- Batanes State College
- Instituto Continental
- Sekolah Menengah Kejuruan Kesehatan Kaltara (Tarakan)
- Colegio de La Inmaculada Concepcion - Cebu